2. High-level interface#
Characterizing neural populations with svGPFA models involves the following steps:
Building an empty model
model = stats.svGPFA.svGPFAModelFactory.SVGPFAModelFactory.buildModel( conditionalDist=stats.svGPFA.svGPFAModelFactory.PointProcess, linkFunction=stats.svGPFA.svGPFAModelFactory.ExponentialLink, embeddingType=stats.svGPFA.svGPFAModelFactory.LinearEmbedding, kernels=kernels)
by specifying a conditional distribution (e.g., point-process,
PointProcess), an embedding type (e.g., linear,LinearEmbedding), a link function (e.g.,exp()) and providing a set of kernels (Kernel).Setting initial parameters
model.setParamsAndData( measurements=spikes_times, initial_params=params["initial_params"], eLLCalculationParams=params["ell_calculation_params"], priorCovRegParam=params["optim_params"]["prior_cov_reg_param"])
Estimating parameters
svEM = stats.svGPFA.svEM.SVEM() lowerBoundHist = svEM.maximize(model=model, measurements=spikeTimes, initialParams=initialParams, quadParams=quadParams, optimParams=optimParams)
by providing a set of measurements,
spikeTimes, initial parameters,initialParams, quadrature parameters,quadParamsand optimisation parameters,optimParams.Assessing goodness-of-fit
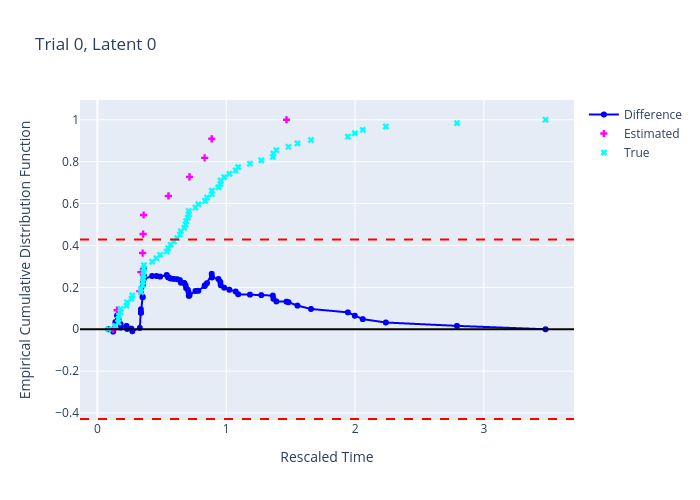
diffECDFsX, diffECDFsY, estECDFx, estECDFy, simECDFx, simECDFy, cb = \ gcnu_common.stats.pointProcesses.tests.\ KSTestTimeRescalingNumericalCorrection(spikes_times=spikes_times_GOF, cif_times=trial_times_GOF, cif_values=cif_values_GOF, gamma=ksTestGamma) fig = svGPFA.plot.plotUtilsPlotly.\ getPlotResKSTestTimeRescalingNumericalCorrection( diffECDFsX=diffECDFsX, diffECDFsY=diffECDFsY, estECDFx=estECDFx, estECDFy=estECDFy, simECDFx=simECDFx, simECDFy=simECDFy, cb=cb, title=title) fig.show()
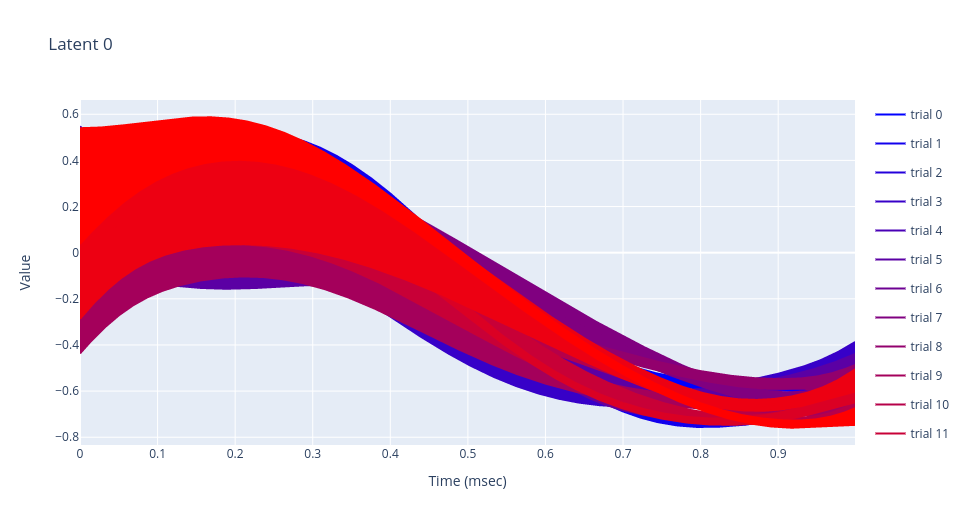
Plotting model’s parameters
fig = svGPFA.plot.plotUtilsPlotly.getPlotLatentAcrossTrials(times=trials_times.numpy(), latentsMeans=testMuK, latentsSTDs=torch.sqrt(testVarK), indPointsLocs=indPointsLocs, latentToPlot=latentToPlot, trials_colors=trials_colors, xlabel="Time (msec)") fig.show()